Publications
Publications and preprints in reversed chronological order.
2026
-
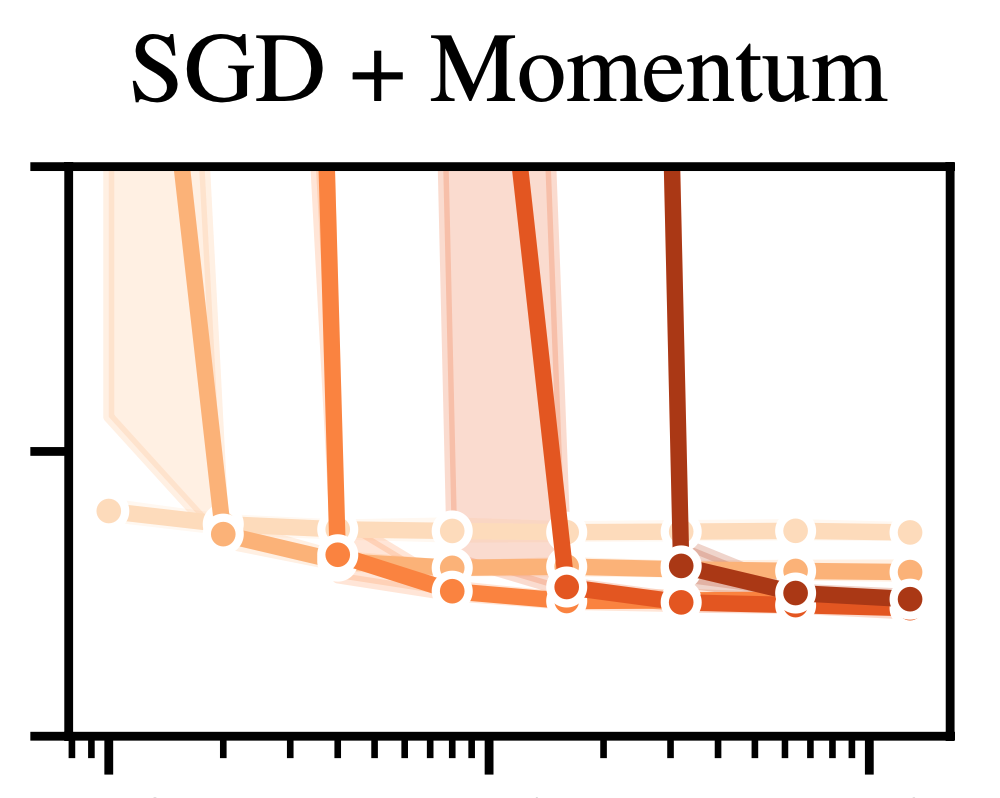 Variational Deep Learning via Implicit RegularizationJonathan Wenger, Beau Coker, Juraj Marusic, and John P. CunninghamInternational Conference on Learning Representations (ICLR), 2026
Variational Deep Learning via Implicit RegularizationJonathan Wenger, Beau Coker, Juraj Marusic, and John P. CunninghamInternational Conference on Learning Representations (ICLR), 2026Modern deep learning models generalize remarkably well in-distribution, despite being overparametrized and trained with little to no explicit regularization. Instead, current theory credits implicit regularization imposed by the choice of architecture, hyperparameters and optimization procedure. However, deep neural networks can be surprisingly non-robust, resulting in overconfident predictions and poor out-of-distribution generalization. Bayesian deep learning addresses this via model averaging, but typically requires significant computational resources as well as carefully elicited priors to avoid overriding the benefits of implicit regularization. Instead, in this work, we propose to regularize variational neural networks solely by relying on the implicit bias of (stochastic) gradient descent. We theoretically characterize this inductive bias in overparametrized linear models as generalized variational inference and demonstrate the importance of the choice of parametrization. Empirically, our approach demonstrates strong in- and out-of-distribution performance without additional hyperparameter tuning and with minimal computational overhead.
@inproceedings{Wenger2025VariationalDeep, title = {Variational Deep Learning via Implicit Regularization}, author = {Wenger, Jonathan and Coker, Beau and Marusic, Juraj and Cunningham, John P.}, booktitle = {International Conference on Learning Representations (ICLR)}, year = {2026}, doi = {10.48550/arXiv.2505.20235}, }
2025
-
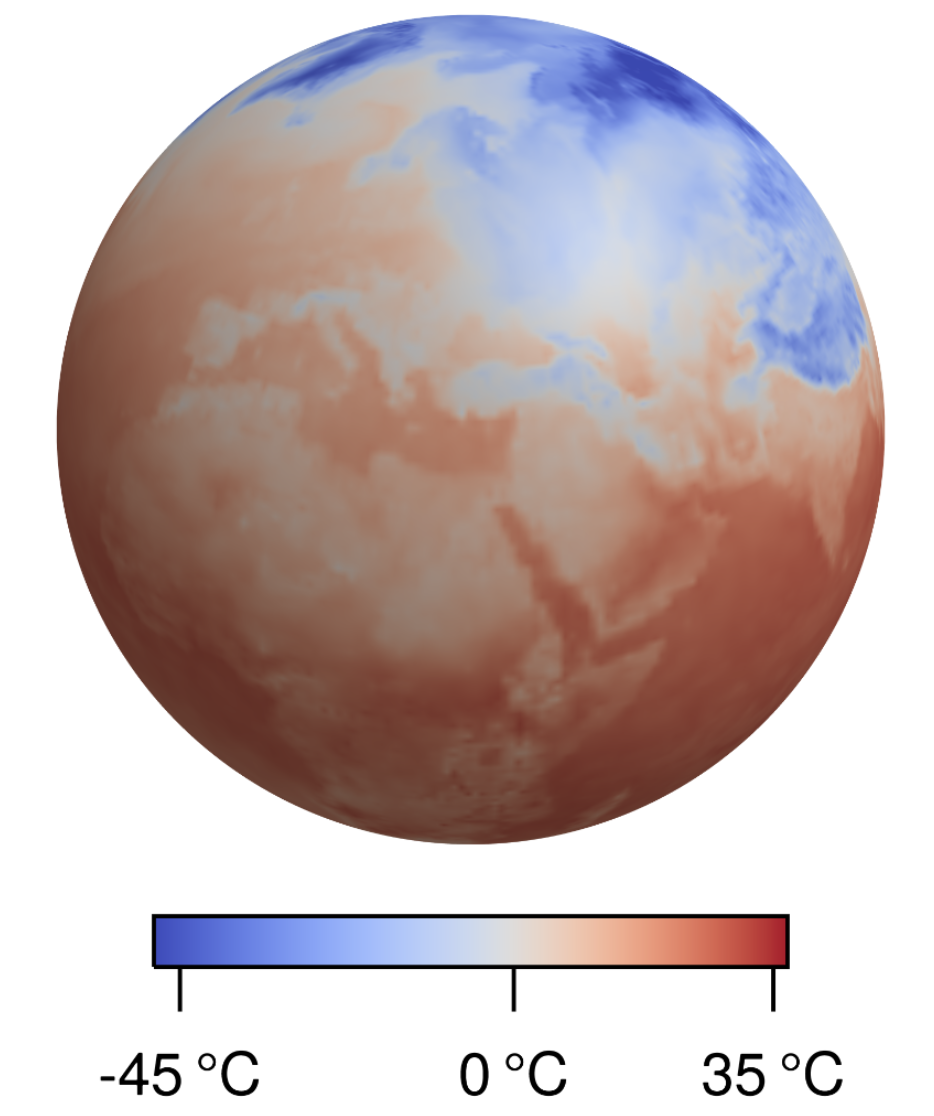 Computation-Aware Kalman Filtering and SmoothingMarvin Pförtner, Jonathan Wenger, Jon Cockayne, and Philipp HennigInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2025
Computation-Aware Kalman Filtering and SmoothingMarvin Pförtner, Jonathan Wenger, Jon Cockayne, and Philipp HennigInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2025Kalman filtering and smoothing are the foundational mechanisms for efficient inference in Gauss–Markov models. However, their time and memory complexities scale prohibitively with the size of the state space. This is particularly problematic in spatiotemporal regression problems, where the state dimension scales with the number of spatial observations. Existing approximate frameworks leverage low-rank approximations of the covariance matrix. But since they do not model the error introduced by the computational approximation, their predictive uncertainty estimates can be overly optimistic. In this work, we propose a probabilistic numerical method for inference in high-dimensional Gauss–Markov models which mitigates these scaling issues. Our matrix-free iterative algorithm leverages GPU acceleration and crucially enables a tunable trade-off between computational cost and predictive uncertainty. Finally, we demonstrate the scalability of our method on a large-scale climate dataset.
@inproceedings{Pfoertner2025ComputationAwareKalman, author = {Pf\"ortner, Marvin and Wenger, Jonathan and Cockayne, Jon and Hennig, Philipp}, title = {Computation-Aware {K}alman Filtering and Smoothing}, year = {2025}, booktitle = {International Conference on Artificial Intelligence and Statistics (AISTATS)}, doi = {10.48550/arxiv.2405.08971}, } -
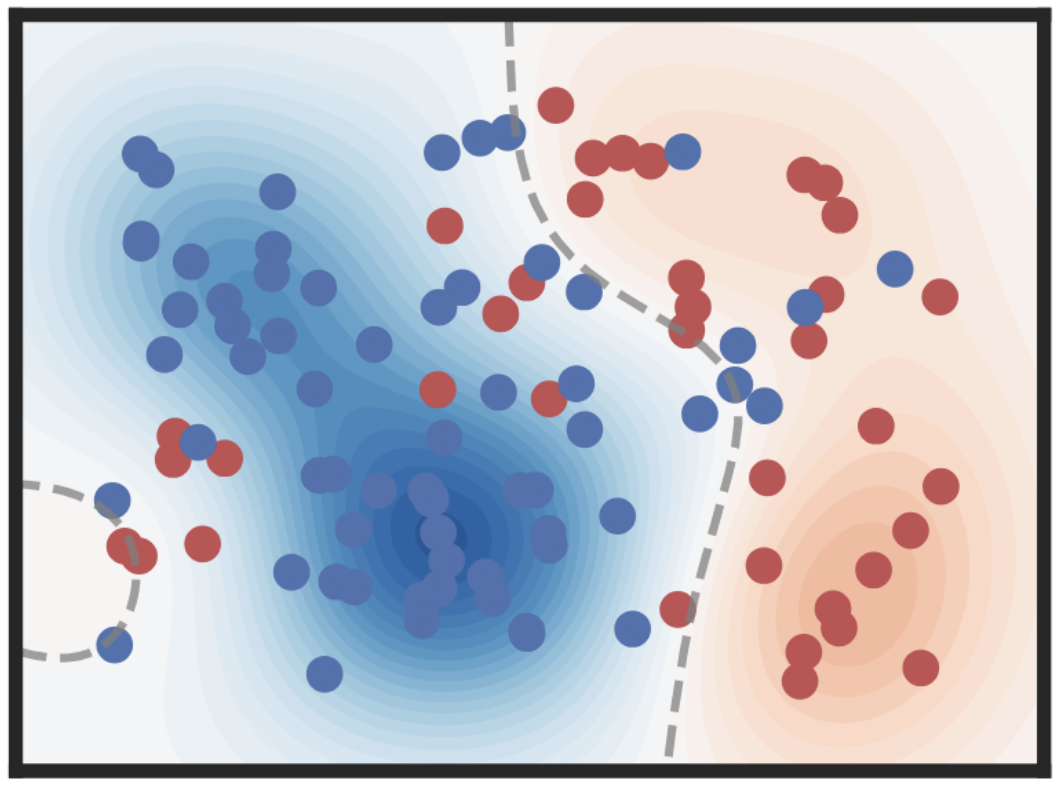 Accelerating Non-Conjugate Gaussian Processes By Trading Off Computation For UncertaintyLukas Tatzel, Jonathan Wenger, Frank Schneider, and Philipp HennigTransactions on Machine Learning Research (TMLR), 2025
Accelerating Non-Conjugate Gaussian Processes By Trading Off Computation For UncertaintyLukas Tatzel, Jonathan Wenger, Frank Schneider, and Philipp HennigTransactions on Machine Learning Research (TMLR), 2025Non-conjugate Gaussian processes (NCGPs) define a flexible probabilistic framework to model categorical, ordinal and continuous data, and are widely used in practice. However, exact inference in NCGPs is prohibitively expensive for large datasets, thus requiring approximations in practice. The approximation error adversely impacts the reliability of the model and is not accounted for in the uncertainty of the prediction. We introduce a family of iterative methods that explicitly model this error. They are uniquely suited to parallel modern computing hardware, efficiently recycle computations, and compress information to reduce both the time and memory requirements for NCGPs. As we demonstrate on large-scale classification problems, our method significantly accelerates posterior inference compared to competitive baselines by trading off reduced computation for increased uncertainty.
@article{Tatzel2025AcceleratingNonConjugate, title = {Accelerating {Non}-{Conjugate} {Gaussian} {Processes} {By} {Trading} {Off} {Computation} {For} {Uncertainty}}, doi = {10.48550/arXiv.2310.20285}, journal = {Transactions on Machine Learning Research (TMLR)}, author = {Tatzel, Lukas and Wenger, Jonathan and Schneider, Frank and Hennig, Philipp}, year = {2025}, }
2024
-
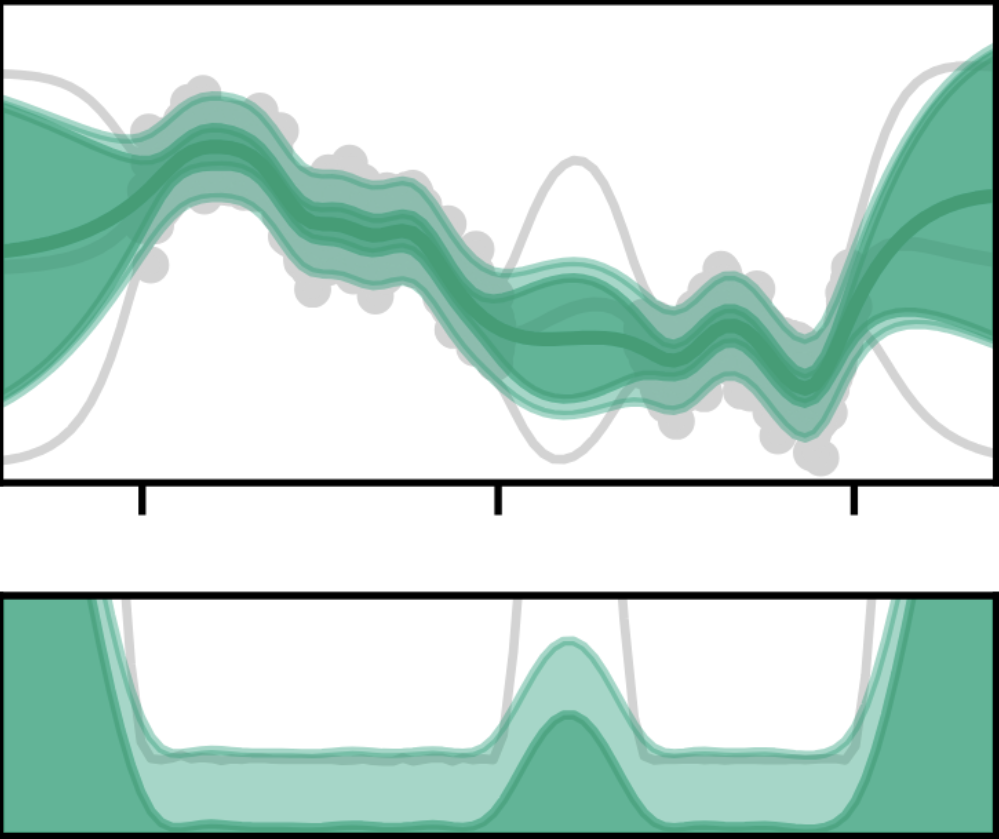 Computation-Aware Gaussian Processes: Model Selection And Linear-Time InferenceJonathan Wenger, Kaiwen Wu, Philipp Hennig, Jacob R. Gardner, Geoff Pleiss, and John P. CunninghamAdvances in Neural Information Processing Systems (NeurIPS), 2024
Computation-Aware Gaussian Processes: Model Selection And Linear-Time InferenceJonathan Wenger, Kaiwen Wu, Philipp Hennig, Jacob R. Gardner, Geoff Pleiss, and John P. CunninghamAdvances in Neural Information Processing Systems (NeurIPS), 2024Model selection in Gaussian processes scales prohibitively with the size of the training dataset, both in time and memory. While many approximations exist, all incur inevitable approximation error. Recent work accounts for this error in the form of computational uncertainty, which enables – at the cost of quadratic complexity – an explicit tradeoff between computation and precision. Here we extend this development to model selection, which requires significant enhancements to the existing approach, including linear-time scaling in the size of the dataset. We propose a novel training loss for hyperparameter optimization and demonstrate empirically that the resulting method can outperform SGPR, CGGP and SVGP, state-of-the-art methods for GP model selection, on medium to large-scale datasets. Our experiments show that model selection for computation-aware GPs trained on 1.8 million data points can be done within a few hours on a single GPU. As a result of this work, Gaussian processes can be trained on large-scale datasets without significantly compromising their ability to quantify uncertainty – a fundamental prerequisite for optimal decision-making.
@inproceedings{Wenger2024ComputationAwareGaussian, title = {Computation-{Aware} {Gaussian} {Processes}: {Model} {Selection} {And} {Linear}-{Time} {Inference}}, author = {Wenger, Jonathan and Wu, Kaiwen and Hennig, Philipp and Gardner, Jacob R. and Pleiss, Geoff and Cunningham, John P.}, booktitle = {Advances in Neural Information Processing Systems (NeurIPS)}, year = {2024}, doi = {10.48550/arXiv.2411.01036}, } -
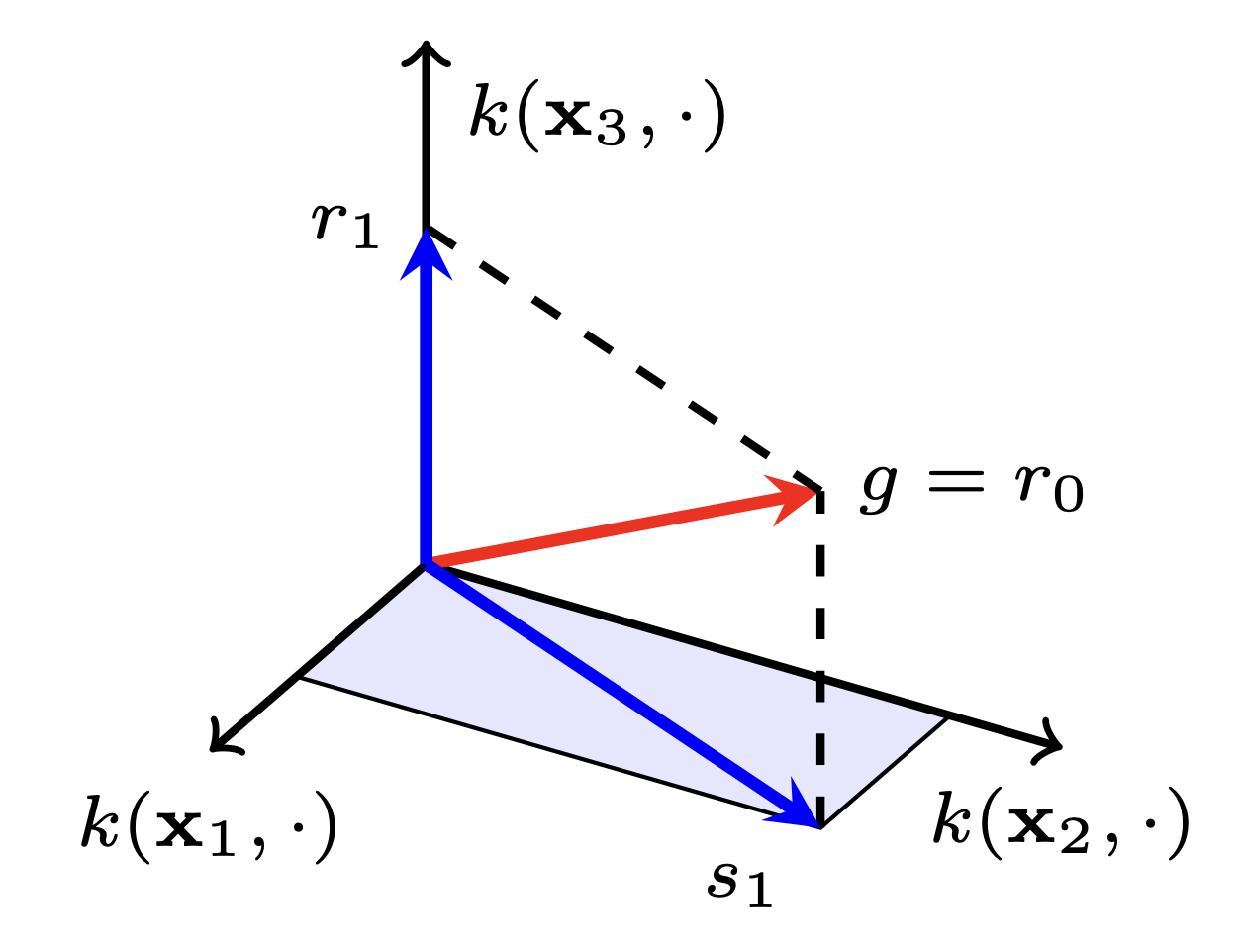 Large-Scale Gaussian Processes via Alternating ProjectionKaiwen Wu, Jonathan Wenger, Haydn Jones, Geoff Pleiss, and Jacob R. GardnerInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2024
Large-Scale Gaussian Processes via Alternating ProjectionKaiwen Wu, Jonathan Wenger, Haydn Jones, Geoff Pleiss, and Jacob R. GardnerInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2024Training and inference in Gaussian processes (GPs) require solving linear systems with n × n kernel matrices. To address the prohibitive O(n^3) time complexity, recent work has employed fast iterative methods, like conjugate gradients (CG). However, as datasets increase in magnitude, the kernel matrices become increasingly ill-conditioned and still require O(n^2) space without partitioning. Thus, while CG increases the size of datasets GPs can be trained on, modern datasets reach scales beyond its applicability. In this work, we propose an iterative method which only accesses subblocks of the kernel matrix, effectively enabling mini-batching. Our algorithm, based on alternating projection, has O(n) per-iteration time and space complexity, solving many of the practical challenges of scaling GPs to very large datasets. Theoretically, we prove the method enjoys linear convergence. Empirically, we demonstrate its fast convergence in practice and robustness to ill-conditioning. On large-scale benchmark datasets with up to four million data points, our approach accelerates GP training and inference by speed-up factors up to 27× and 72×, respectively, compared to CG.
@inproceedings{Wu2024LargeScaleGaussian, title = {Large-{Scale} {Gaussian} {Processes} via {Alternating} {Projection}}, author = {Wu, Kaiwen and Wenger, Jonathan and Jones, Haydn and Pleiss, Geoff and Gardner, Jacob R.}, booktitle = {International Conference on Artificial Intelligence and Statistics (AISTATS)}, year = {2024}, doi = {10.48550/arXiv.2310.17137}, }
2023
-
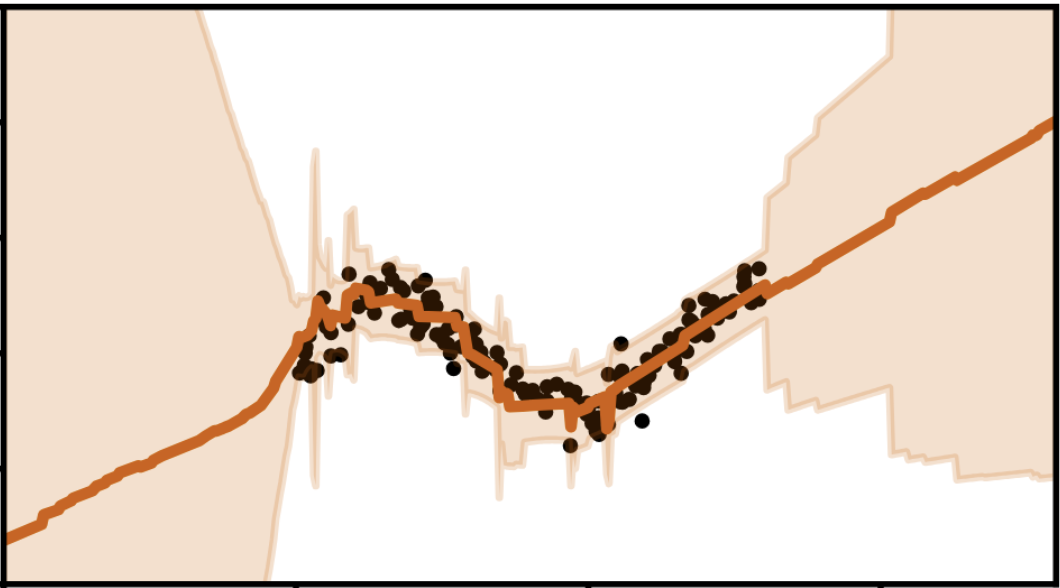 On the Disconnect Between Theory and Practice of Neural Networks: Limits of the Neural Tangent Kernel PerspectiveJonathan Wenger, Felix Dangel, and Agustinus Kristiadi2023
On the Disconnect Between Theory and Practice of Neural Networks: Limits of the Neural Tangent Kernel PerspectiveJonathan Wenger, Felix Dangel, and Agustinus Kristiadi2023The neural tangent kernel (NTK) has garnered significant attention as a theoretical framework for describing the behavior of large-scale neural networks. Kernel methods are theoretically well-understood and as a result enjoy algorithmic benefits, which can be demonstrated to hold in wide synthetic neural network architectures. These advantages include faster optimization, reliable uncertainty quantification and improved continual learning. However, current results quantifying the rate of convergence to the kernel regime suggest that exploiting these benefits requires architectures that are orders of magnitude wider than they are deep. This assumption raises concerns that architectures used in practice do not exhibit behaviors as predicted by the NTK. Here, we supplement previous work on the NTK by empirically investigating whether the limiting regime predicts practically relevant behavior of large-width architectures. Our results demonstrate that this is not the case across multiple domains. This observed disconnect between theory and practice further calls into question to what degree NTK theory should inform architectural and algorithmic choices.
@misc{Wenger2023DisconnectTheory, title = {On the {Disconnect} {Between} {Theory} and {Practice} of {Neural} {Networks}: Limits of the Neural Tangent Kernel Perspective}, author = {Wenger, Jonathan and Dangel, Felix and Kristiadi, Agustinus}, howpublished = {arXiv}, year = {2023}, doi = {10.48550/arXiv.2310.00137}, } -
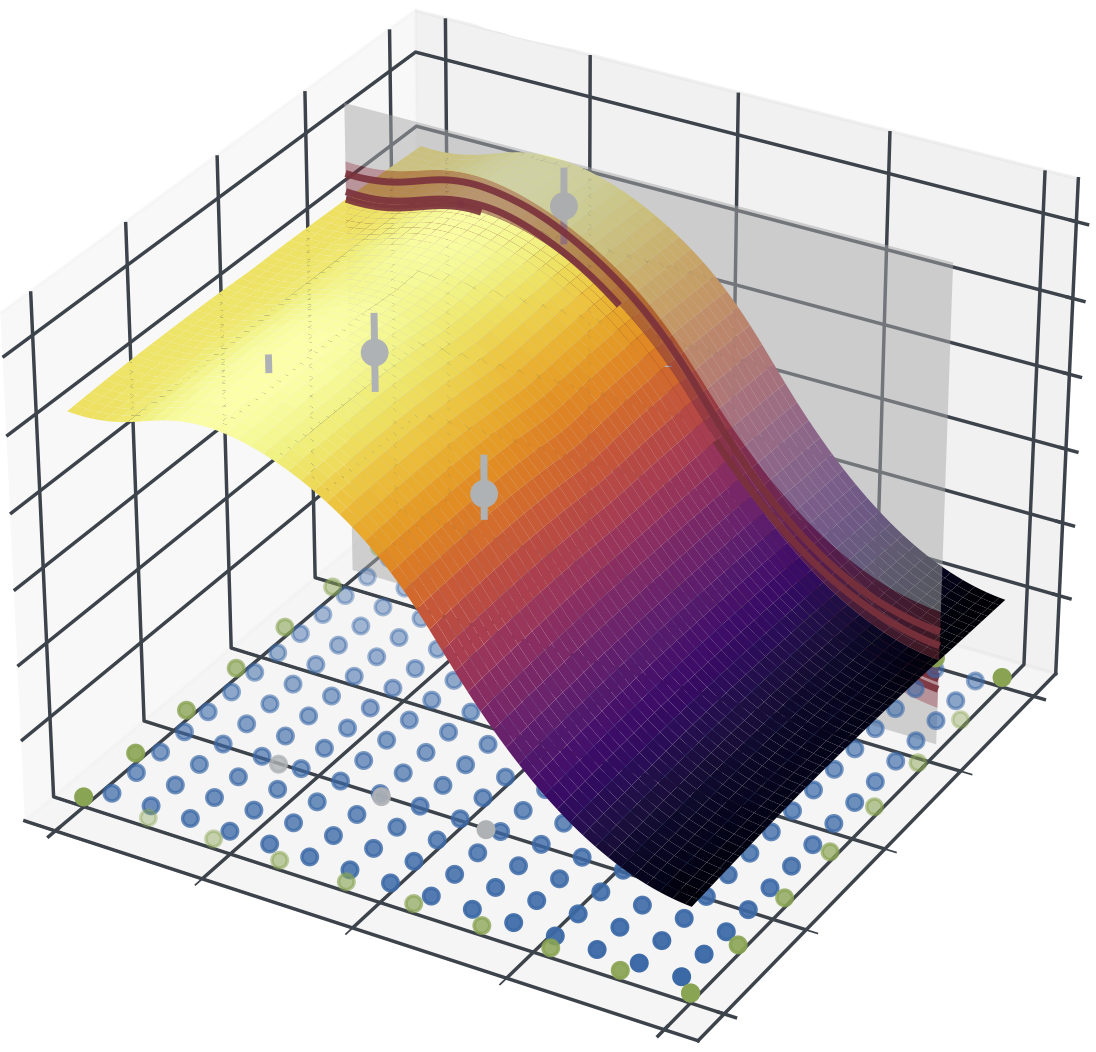 Physics-Informed Gaussian Process Regression Generalizes Linear PDE SolversMarvin Pförtner, Ingo Steinwart, Philipp Hennig, and Jonathan Wenger2023
Physics-Informed Gaussian Process Regression Generalizes Linear PDE SolversMarvin Pförtner, Ingo Steinwart, Philipp Hennig, and Jonathan Wenger2023Linear partial differential equations (PDEs) are an important, widely applied class of mechanistic models, describing physical processes such as heat transfer, electromagnetism, and wave propagation. In practice, specialized numerical methods based on discretization are used to solve PDEs. They generally use an estimate of the unknown model parameters and, if available, physical measurements for initialization. Such solvers are often embedded into larger scientific models with a downstream application and thus error quantification plays a key role. However, by ignoring parameter and measurement uncertainty, classical PDE solvers may fail to produce consistent estimates of their inherent approximation error. In this work, we approach this problem in a principled fashion by interpreting solving linear PDEs as physics-informed Gaussian process (GP) regression. Our framework is based on a key generalization of the Gaussian process inference theorem to observations made via an arbitrary bounded linear operator. Crucially, this probabilistic viewpoint allows to (1) quantify the inherent discretization error; (2) propagate uncertainty about the model parameters to the solution; and (3) condition on noisy measurements. Demonstrating the strength of this formulation, we prove that it strictly generalizes methods of weighted residuals, a central class of PDE solvers including collocation, finite volume, pseudospectral, and (generalized) Galerkin methods such as finite element and spectral methods. This class can thus be directly equipped with a structured error estimate. In summary, our results enable the seamless integration of mechanistic models as modular building blocks into probabilistic models by blurring the boundaries between numerical analysis and Bayesian inference.
@misc{Pfortner2023PhysicsInformedGaussian, title = {Physics-{Informed} {Gaussian} {Process} {Regression} {Generalizes} {Linear} {PDE} {Solvers}}, author = {Pförtner, Marvin and Steinwart, Ingo and Hennig, Philipp and Wenger, Jonathan}, howpublished = {arXiv}, year = {2023}, doi = {10.48550/arXiv.2212.12474}, }
2022
-
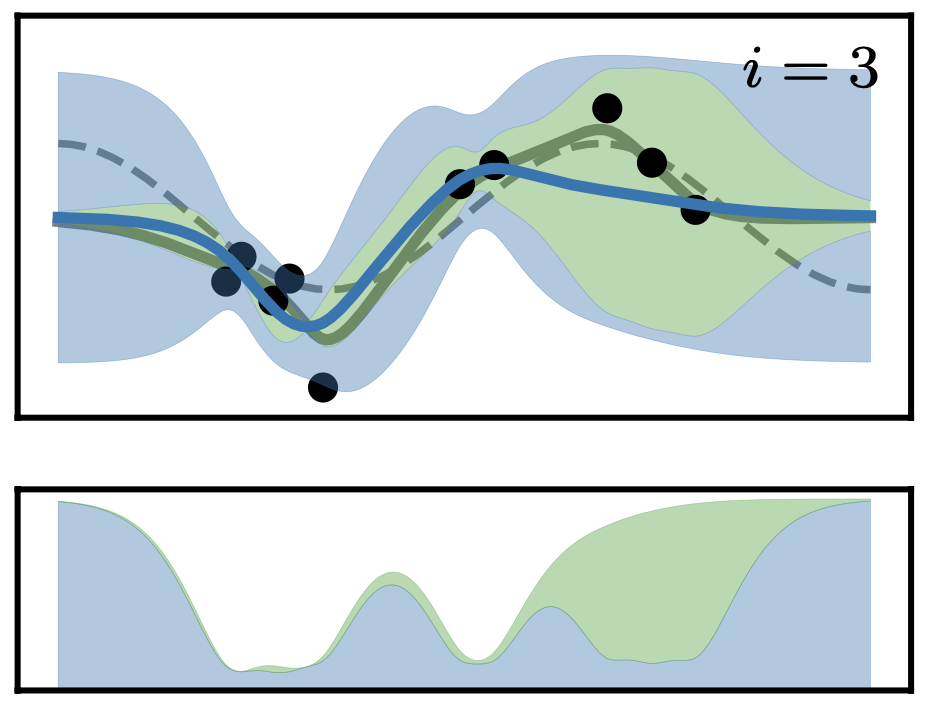 Posterior and Computational Uncertainty in Gaussian ProcessesJonathan Wenger, Geoff Pleiss, Marvin Pförtner, Philipp Hennig, and John P. CunninghamAdvances in Neural Information Processing Systems (NeurIPS), 2022
Posterior and Computational Uncertainty in Gaussian ProcessesJonathan Wenger, Geoff Pleiss, Marvin Pförtner, Philipp Hennig, and John P. CunninghamAdvances in Neural Information Processing Systems (NeurIPS), 2022Gaussian processes scale prohibitively with the size of the dataset. In response, many approximation methods have been developed, which inevitably introduce approximation error. This additional source of uncertainty, due to limited computation, is entirely ignored when using the approximate posterior. Therefore in practice, GP models are often as much about the approximation method as they are about the data. Here, we develop a new class of methods that provides consistent estimation of the combined uncertainty arising from both the finite number of data observed and the finite amount of computation expended. The most common GP approximations map to an instance in this class, such as methods based on the Cholesky factorization, conjugate gradients, and inducing points. For any method in this class, we prove (i) convergence of its posterior mean in the associated RKHS, (ii) decomposability of its combined posterior covariance into mathematical and computational covariances, and (iii) that the combined variance is a tight worst-case bound for the squared error between the method’s posterior mean and the latent function. Finally, we empirically demonstrate the consequences of ignoring computational uncertainty and show how implicitly modeling it improves generalization performance on benchmark datasets.
@inproceedings{Wenger2022PosteriorComputational, title = {Posterior and Computational Uncertainty in {Gaussian} Processes}, author = {Wenger, Jonathan and Pleiss, Geoff and Pf{\"o}rtner, Marvin and Hennig, Philipp and Cunningham, John P.}, booktitle = {Advances in Neural Information Processing Systems (NeurIPS)}, year = {2022}, doi = {10.48550/arXiv.2205.15449}, } -
 Preconditioning for Scalable Gaussian Process Hyperparameter OptimizationJonathan Wenger, Geoff Pleiss, Philipp Hennig, John P. Cunningham, and Jacob R. GardnerInternational Conference on Machine Learning (ICML), 2022
Preconditioning for Scalable Gaussian Process Hyperparameter OptimizationJonathan Wenger, Geoff Pleiss, Philipp Hennig, John P. Cunningham, and Jacob R. GardnerInternational Conference on Machine Learning (ICML), 2022Gaussian process hyperparameter optimization requires linear solves with, and log-determinants of, large kernel matrices. Iterative numerical techniques are becoming popular to scale to larger datasets, relying on the conjugate gradient method (CG) for the linear solves and stochastic trace estimation for the log-determinant. This work introduces new algorithmic and theoretical insights for preconditioning these computations. While preconditioning is well understood in the context of CG, we demonstrate that it can also accelerate convergence and reduce variance of the estimates for the log-determinant and its derivative. We prove general probabilistic error bounds for the preconditioned computation of the log-determinant, log-marginal likelihood and its derivatives. Additionally, we derive specific rates for a range of kernel-preconditioner combinations, showing that up to exponential convergence can be achieved. Our theoretical results enable provably efficient optimization of kernel hyperparameters, which we validate empirically on large-scale benchmark problems. There our approach accelerates training by up to an order of magnitude.
@inproceedings{Wenger2022PreconditioningScalable, title = {Preconditioning for Scalable {G}aussian Process Hyperparameter Optimization}, author = {Wenger, Jonathan and Pleiss, Geoff and Hennig, Philipp and Cunningham, John P. and Gardner, Jacob R.}, booktitle = {International Conference on Machine Learning (ICML)}, year = {2022}, doi = {10.48550/arXiv.2107.00243}, }
2021
-
 ProbNum: Probabilistic Numerics in PythonJonathan Wenger, Nicholas Krämer, Marvin Pförtner, Jonathan Schmidt, Nathanael Bosch, Nina Effenberger, Johannes Zenn, Alexandra Gessner, Toni Karvonen, François-Xavier Briol, Maren Mahsereci, and Philipp Hennig2021
ProbNum: Probabilistic Numerics in PythonJonathan Wenger, Nicholas Krämer, Marvin Pförtner, Jonathan Schmidt, Nathanael Bosch, Nina Effenberger, Johannes Zenn, Alexandra Gessner, Toni Karvonen, François-Xavier Briol, Maren Mahsereci, and Philipp Hennig2021Probabilistic numerical methods (PNMs) solve numerical problems via probabilistic inference. They have been developed for linear algebra, optimization, integration and differential equation simulation. PNMs naturally incorporate prior information about a problem and quantify uncertainty due to finite computational resources as well as stochastic input. In this paper, we present ProbNum: a Python library providing state-of-the-art probabilistic numerical solvers. ProbNum enables custom composition of PNMs for specific problem classes via a modular design as well as wrappers for off-the-shelf use. Tutorials, documentation, developer guides and benchmarks are available online at https://probnum.readthedocs.io/.
@misc{Wenger2021ProbNumProbabilistic, title = {{ProbNum}: {Probabilistic} {Numerics} in {Python}}, author = {Wenger, Jonathan and Krämer, Nicholas and Pförtner, Marvin and Schmidt, Jonathan and Bosch, Nathanael and Effenberger, Nina and Zenn, Johannes and Gessner, Alexandra and Karvonen, Toni and Briol, François-Xavier and Mahsereci, Maren and Hennig, Philipp}, year = {2021}, howpublished = {arXiv}, }
2020
-
 Probabilistic Linear Solvers for Machine LearningJonathan Wenger, and Philipp HennigAdvances in Neural Information Processing Systems (NeurIPS), 2020
Probabilistic Linear Solvers for Machine LearningJonathan Wenger, and Philipp HennigAdvances in Neural Information Processing Systems (NeurIPS), 2020Linear systems are the bedrock of virtually all numerical computation. Machine learning poses specific challenges for the solution of such systems due to their scale, characteristic structure, stochasticity and the central role of uncertainty in the field. Unifying earlier work we propose a class of probabilistic linear solvers which jointly infer the matrix, its inverse and the solution from matrix-vector product observations. This class emerges from a fundamental set of desiderata which constrains the space of possible algorithms and recovers the method of conjugate gradients under certain conditions. We demonstrate how to incorporate prior spectral information in order to calibrate uncertainty and experimentally showcase the potential of such solvers for machine learning.
@inproceedings{Wenger2020ProbabilisticLinear, title = {Probabilistic {Linear} {Solvers} for {Machine} {Learning}}, author = {Wenger, Jonathan and Hennig, Philipp}, booktitle = {Advances in Neural Information Processing Systems (NeurIPS)}, year = {2020}, doi = {10.48550/arXiv.2010.09691}, } -
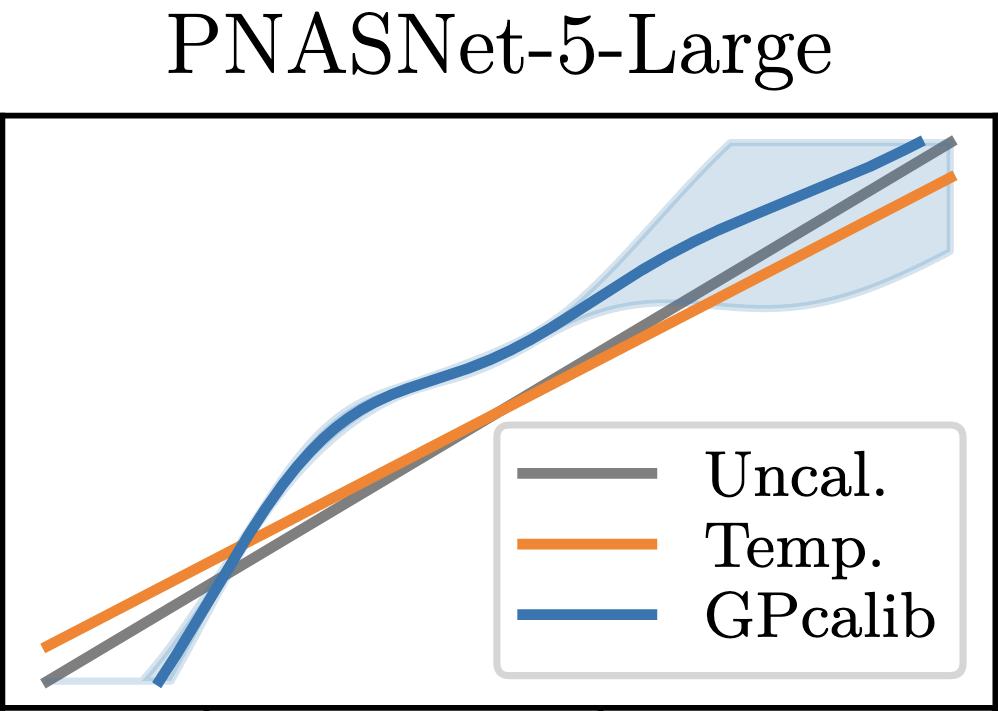 Non-Parametric Calibration for ClassificationJonathan Wenger, Hedvig Kjellström, and Rudolph TriebelInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2020
Non-Parametric Calibration for ClassificationJonathan Wenger, Hedvig Kjellström, and Rudolph TriebelInternational Conference on Artificial Intelligence and Statistics (AISTATS), 2020Many applications of classification methods not only require high accuracy but also reliable estimation of predictive uncertainty. However, while many current classification frameworks, in particular deep neural networks, achieve high accuracy, they tend to incorrectly estimate uncertainty. In this paper, we propose a method that adjusts the confidence estimates of a general classifier such that they approach the probability of classifying correctly. In contrast to existing approaches, our calibration method employs a non-parametric representation using a latent Gaussian process, and is specifically designed for multi-class classification. It can be applied to any classifier that outputs confidence estimates and is not limited to neural networks. We also provide a theoretical analysis regarding the over- and underconfidence of a classifier and its relationship to calibration, as well as an empirical outlook for calibrated active learning. In experiments we show the universally strong performance of our method across different classifiers and benchmark data sets, in particular for state-of-the art neural network architectures.
@inproceedings{Wenger2020NonParametricCalibration, title = {Non-{Parametric} {Calibration} for {Classification}}, author = {Wenger, Jonathan and Kjellström, Hedvig and Triebel, Rudolph}, booktitle = {International Conference on Artificial Intelligence and Statistics (AISTATS)}, year = {2020}, doi = {10.48550/arXiv.1906.04933}, }